GWAS became possible about 10 years ago as the result of several scientific advances. Since then, GWAS has continually developed as a primary method for identification of disease susceptibility genes in humans and other organisms. At Golden Helix we are proud of our history in supporting GWAS analysis from its inception. Our software was used to analyze whole-genome data from… Read more »
Earlier this spring we announced that Meta-Analysis was coming to SVS very soon. Now, I am pleased to announce that it is available in the latest release of SVS (version 8.4.0). Meta-Analysis takes the results of two or more GWAS studies for multiple SNPs or markers, and standard meta-analysis statistics are then performed on each SNP and the results compiled into… Read more »
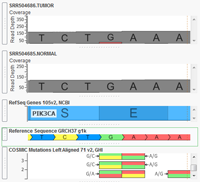
VarSeq now supports analysis of paired Tumor/Normal samples! Tumor/Normal support has been one of the most common feature requests for VarSeq since it was launched late last year, and we are excited to make this functionality available to all of our VarSeq users in the latest update (version 1.1.4). VarSeq is a powerful platform for annotation and filtering of DNA… Read more »
One of the lesser known functions in SVS allows the user to create Venn diagrams comparing variants found in multiple spreadsheets. These different spreadsheets could come from individuals samples, a case vs. control group or several variant databases. It is a helpful tool for visually comparing different variants. Start by creating spreadsheets for each group/samples you’d like to compare. The… Read more »
It is already almost halfway through 2015, and June has been especially busy as far as customer publication goes. We wanted to pass on the articles to you and congratulate our customers on their success! Elena Gusareva and Kristel Van Steen of the University of Liege and colleagues recently published A cautionary note on the impact of protocol changes for genome-wide… Read more »
The support team at Golden Helix is always on-hand to help with your SVS and VarSeq needs. We get some questions more often than others, and this blog will answer some of the most common questions we’ve been seeing lately regarding VarSeq. A common question we receive is if data can be filtered from a locally kept set of variants… Read more »
There are many approaches that one might use to define a variant as potentially deleterious. For example, we often see analysis workflows based on rare, non-synonymous variants, perhaps incorporating additional annotation sources that capture known or predicted consequences of coding variants. Annotations for coding regions of the genome are relatively abundant and familiar to genome scientists. We are comfortable in… Read more »
This year the MAGES symposium has a new name, the Symposium on Advances in Genomics, Epidemiology and Statistics or SAGES! The NIH has also been added to the sponsor list and we’re excited to have the support for this informative symposium! Additionally, we were treated to the addition of a new poster session to accompany the fantastic speakers. The trend… Read more »
Today we wanted to share a recent client case study to demonstrate how VarSeq has been implemented in a CLIA certified clinical laboratory. Please feel free to contact us if you have questions or if you would like to discuss VarSeq further at info@goldenhelix.com. It is standard practice for newborns to be screened for genetic diseases before leaving the hospital to… Read more »
Today at Golden Helix, we are proud to announce our collaboration with MedGenome through an integration of OncoMD into our VarSeq software. Now VarSeq’s streamlined process of annotating and filtering variants will offer an added dimension. OncoMD is a comprehensive knowledge base of cancer-specific genetic alterations, and by incorporating it into VarSeq, users can access to 2 million plus annotated… Read more »
There’s a strong desire in the genetics community for a set of canonical transcripts. It’s a completely understandable and reasonable thing to want since it would simplify many aspects of analysis and especially the downstream communicating and reporting of variants. Unfortunately, biology isn’t so tidy as to provide a clear answer for which transcript is the important one. Consequently, there… Read more »
Several of our customers have published recently, using our SVS and VarSeq software, and we love sharing their work with you. Congrats to all! Sebastian Mucha and Joanne Conington of Scotland’s Rural College along with colleagues collaboratively published Genome-wide association study of footrot in Texel sheep in Genetics Selection Evolution which used GWAS to investigate links between ovine footrot scores and molecular polymorphisms… Read more »
Meta-analysis is an important tool to have in the bioinformatics toolbox. The numbers alone speak for themselves. It is the fourth most requested feature for SVS, and a simple google scholar search for 2014 and 2015 find 17,300 results for genetics + meta-analysis. There are several meta-analysis utilities out there that will take results from studies and perform the meta-analysis…. Read more »
The SNP and Variation Suite (SVS) software currently supports three methods for genomic prediction: Genomic Best Linear Unbiased Predictors (GBLUP), Bayes C and Bayes C-pi. We have discussed these methods extensively in previous blogs and webcast events. Although there are extensive applications for these methods, they are primarily used for trait selection in agricultural genetics. Each method can be used… Read more »
Our VarSeq as a Clinical Platform webcast last week highlighted some recent updates in VarSeq that support gene panel screenings and rare variant diagnostics. The webcast generated some good questions, and I wanted to share them with you. If the questions below spark new questions or need clarification, feel free to get in touch with us at info@goldenhelix.com. Question: Should… Read more »
Over 650 GenomeBrowse licenses have been registered and downloaded since the beginning of 2015, and with so many people enjoying the utility of this freeware program, I wanted to showcase some advanced tips and tricks so you can get more out of GenomeBrowse! Under the Controls panel, when you’re clicked inside a data plot, there is a “Filter” tab. This… Read more »
Some of our customers have recently published using our SVS and VarSeq software in their studies. We wanted to share their work and congratulate everyone on their success! Maria Skerenova at the Institute of Clinical Biochemistry and her colleagues published Genetic variants in interleukin 7 receptor α chain (IL-7Ra) are associated with multiple sclerosis risk and disability progression in Central European… Read more »
This week, Dr. Jeffery Moore presented a webcast on the Molecular Sciences Made Personal. The webcast delved into Dr. Moore’s attempts to transform how they teach chemistry at the University of Illinois and demonstrated how he uses VarSeq with his students to examine exome data. The following are the questions asked by the attendees. Please feel free to reach out… Read more »
Over the last year our blog has seen a boom in visits and of course, I became curious. What brings people to “Our 2 SNPs…”? So, I decided to take a look at the blog posts that our community find the most intriguing. Here are my findings: Comparing BEAGLE, IMPUTE2, and Minimac Imputation Methods for Accuracy, Computation Time, and Memory… Read more »
I am constantly on the lookout for fun or interesting datasets to analyze in SVS or VarSeq and recently came across a study looking into inherited cardiac conduction disease in an extended family (Lai et al. 2013). The researchers sequenced the exomes from five family members including three affected siblings and their unaffected mother and an unaffected child of one… Read more »