About Darby Kammeraad
Darby Kammeraad is the Director of Field Application Services at Golden Helix, joining the team in April of 2017. Darby graduated in 2016 with a master’s degree in Plant Sciences from Montana State University, where he also received his bachelor’s degree in Plant Biotechnology. Darby works on customer support and training. When not in the office, Darby is learning how to play guitar, hunting, fishing, snowboarding, traveling or working on a new recipe in the kitchen.
The support team at Golden Helix is always here to help with your SVS and VarSeq needs. Often, we receive some excellent questions that should be shared with the rest of our users. This blog will answer some common questions we’ve been seeing lately regarding VarSeq CNV. I’ve noticed there is a version 2 of the CNV caller on Targeted Regions Algorithm, how has… Read more »
VarSeq enables breakthrough discoveries in cancer diagnostics by supporting gene panel testing and whole exome and genome analysis. We wanted to share our Cancer Gene Panel tutorial which covers a basic gene panel workflow with an emphasis on adding, modifying and manipulating filter chains. This tutorial will start with creating a new project from an empty project template, importing data, creating… Read more »
The new VSReports tutorial covers a basic VSReports workflow with an emphasis on understanding and exploring report customizations. This tutorial requires an active VarSeq license with the the VSReport feature included. You can go to Discover VarSeq or email info@goldenhelix.com to request an evaluation license with the VSReports functionality included. VS Reports provides the ability to generate clinical-grade reports. This feature enables VarSeq… Read more »
The new VSWarehouse Tutorial covers the basic VSWarehouse workflow.This tutorial focuses on connecting to a VSWarehouse instance from VarSeq, adding an existing VSWarehouse project as an annotation source and using reports and assessment catalogs hosted on VSWarehouse. This workflow requires an active VarSeq license with the VSWarehouse feature included. You can go to Discover VarSeq or email info@goldenhelix.com to request an… Read more »
Earlier this year we released our own optimized and integrated BEAGLE implementation for SVS based on the BEAGLE 4.1 and optionally 4.0 algorithms. One of the commonly requested features since that released was to expand the algorithm implementation to be considerate of the parent-offspring relationship between samples to inform and improve the accuracy of the haplotype phasing. With this information,… Read more »
Ever since the MacArther Lab announced the new gnomAD browser at last year’s ASHG conference, we have had many requests from our customers to make this new variant frequency source available within both VarSeq and SVS. This new dataset includes variants obtained from 123,136 exome sequences and 15,496 whole-genome sequences. In comparison to the original ExAC dataset which contained exomes… Read more »
In the new Genotype Imputation tool that is coming soon to SVS, allele encoding is an important part of matching data between the target and the reference panels. If the same platform provider is being used, then A/B encoding can be used. However, it’s better to use the Reference/Alternate allele encoding associated with AGCT format to ensure accuracy. If an… Read more »
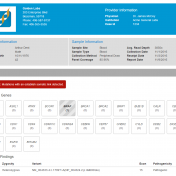
Clinical labs need to be able to process samples down to a short list of variants and publish a professional report. VSReports helps scientists and clinicians alike create timely, actionable reports that can improve clinical decision making and streamline patient care by seamlessly incorporating the results of tertiary analysis into a customizable clinical report. To include the VSReports functionality in… Read more »
Variant interpretation is an integral part of any workflow that results in some decisions being made about the validity and suspected functional impact of a variant in a given sample and their presenting phenotypes. The VarSeq Assessment Catalog functionality is designed to assist the VarSeq user in streamlining this process. To include this functionality in your workflow, you will first… Read more »
True to its nature, VarSeq offers multiple data export options. You can export result tables from VarSeq to Text, VCF, a VarSeq annotation file and most importantly an XLSX (Excel) File. VarSeq’s Excel export options provide a lot of flexibility in the information that is exported and preserve the formatting of data during the export process from VarSeq to Excel. This… Read more »
The new Annotate and Filter algorithm is now available with the release of SVS 8.6.0, see the release notes for full details on all new and updated features. To access this new functionality, you simply need to update your SVS installation to the new version. The update can be done by clicking the Update Available link at the bottom of… Read more »
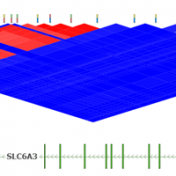
Pruning your data based on Linkage Disequilibrium (LD) values is an important quality assurance step for GWAS analysis. In particular, some tests such as Identity by Descent Estimation (IBD), Inbreeding Coefficient Estimation (f) and Principal Component Analysis (PCA) will obtain better results if the markers used are not in linkage disequilibrium with each other. Therefore, Golden Helix’s SVS provides the… Read more »
Question: Now that I’ve added annotation sources for my sample, filtered down to a list of interesting variants, flagged those variants and generated a clinical report, can I save or copy the annotation sources and filters for use on another sample? Short Answer: Yes! Long Answer: VarSeq was created with ease and efficiency in mind. In VarSeq, once you’ve defined… Read more »
ClinVar is one of our most used annotations sources for a variety of workflows. It is also the public annotation source that is updated most frequently of all the sources currently supported in VarSeq. ClinVar provides new versions of their database once a month in several formats (XML, VCF, TXT). We use custom Python scripts to convert the provided VCF… Read more »
Whole exome sequencing workflows using SNP & Variation Suite (SVS) was presented in a recent webcast, by Dr. Robert Hamilton from the Hospital for Sick Kids. In particular, he performed some filtering on his data to look for only heterozygous variants in his sample of interest, removed variants with allele frequency less than 0.005% based off of the ExAC Variant Frequency… Read more »
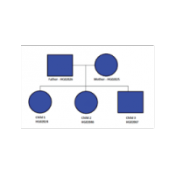
During the webcast yesterday, I demonstrated a few ways of customizing de Novo Candidate and Compound Heterozygous Candidate workflows to consider family structure that was slightly different from the default trio workflow. The families included additional affected and unaffected siblings added to a trio as well as looking at what could be done if there were only two affected siblings… Read more »
Solving the Eigenvalue Decomposition Problem for Large Sample Sizes Since our introduction of the mixed model methods in SVS, along with GBLUP, we have been very pleased to see it used by a number of customers working with human and agri-genomic data. As these customers have grown their genomics programs, the number of samples they have for a given analysis… Read more »
Submit directly to N-of-One from VarSeq If you or your lab uses N-of-One solutions for clinical annotations, here’s some good news: You can now submit directly to N-of-One from VarSeq! N-of-One’s set of preferred transcripts may differ from those outputted by our algorithms in VarSeq, so our solution was built with that in mind. Our slick, easy to use, and… Read more »
Clinical reports come in all shapes, sizes and flavors. With that in mind, our clinical reporting interface VSReports was built to be highly customizable and flexible. With a little Javascript and HTML know-how, your clinical reports can be customized to meet the needs and goals of your lab. With a little Javascript and HTML know how, you can customize yours as… Read more »
Did you know you can analyze your Genotyping by Sequencing (GBS) data in SVS? Well you can! You can combine tools for both GWAS quality control and analysis with tools for NGS data analysis to either identify SNPs in your dataset or to identify differences between populations or sub-species. If your species has a reference sequence or even if you… Read more »