In part one of this series, we discussed how the ACMG Classifier can be implemented in your filter chain to support a best practice workflow. To continue our discussion on best practices of VSClinical, this blog will shed light on other attributes of VSClinical that can add support to your evaluation. Specifically, we will explore how VSClinical can help users understand the ACMG criterion and alleviate the complexity of searching for genetic evidence.
To begin, VSClinical provides an Evidence section that gives insight into the ACMG criteria. The link “See further discussion on PP3/BP4” in the evidence section, shown below, connects you to a variety of useful information. This information applies to all criteria and includes a summary of the ACMG criteria as well as why the criteria were auto-selected by VSClinical. This feature further provides a description from the ACMG guidelines, Clarification from
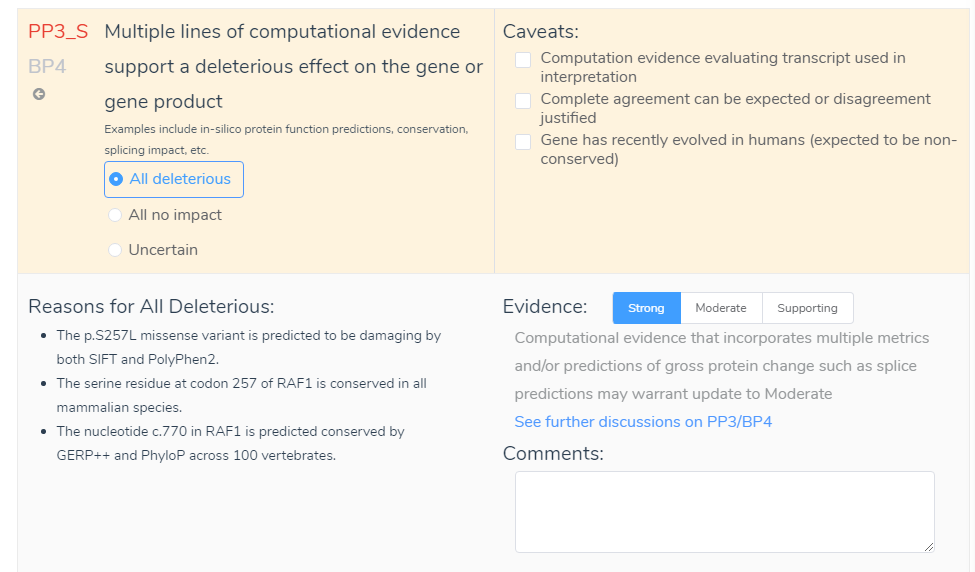

Next is the Comment section in the ACMG criteria. Although not considered to provide available variant evidence, the comment section can serve as a repository for user validations. For example, say the Evidence was changed from Supporting to Strong. Although this decision will be recorded in the Evidence List, you may also want to include information in the comment section. This may include, for example, that the variant influenced all functional prediction (SIFT, PolyPhen, GERP and PhyloP) and splice site prediction (GeneSplicer, MaxEntScan, NN and PWM) algorithms. This comment would then be stored in the interpretation section for the variant and would provide specific reasonings for the evaluation. This also leads to the point that the comment section can serve as a source to view previous user notes. If multiple people use the same assessment catalog, they will be able to see previous variant evaluations which will include the Evidence and Comments that were recorded. Lastly, as shown above, all information from both the Evidence List and Comments can be added to the overall interpretation by hovering over the blue add icon and selecting Add Text to My Interpretation.
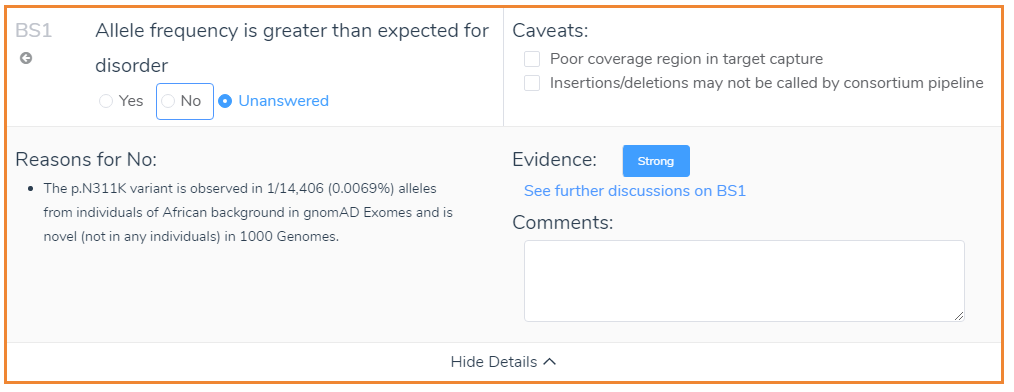
In addition to the Evidence and Comment sections, it is also worth mentioning the Caveats. The Caveats of VSClinical are criterion specific and highlight discrepancies that should be considered when interpreting genetic evidence. A good example is the caveats that appear when looking at the frequency of the variant in the population catalogs: gnomAD Exomes and 1000 Genomes. The caveats that apply for BS1 are shown below and refer to the quality of coverage and the type of variant. Although some caveats may be hard to interpret, VSClinical offers multiple avenues to deep dive into the genetic evidence.
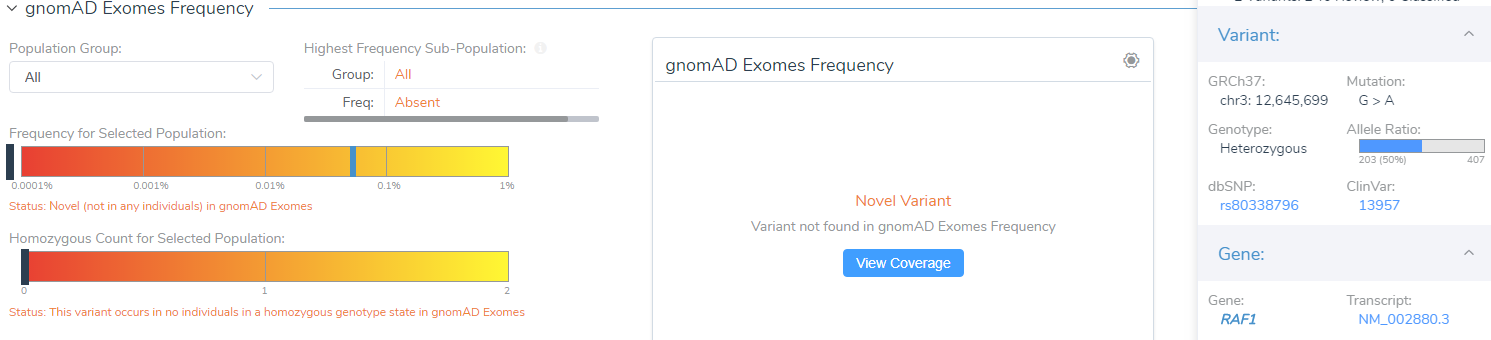
For instance, VSClinical provides a link to visualize the coverage information for the variant displayed in either gnomAD Exomes or 1000 Genomes, shown below. This link will display the coverage information based on exomes and genomes and can be a useful resource when gnomAD Exomes and 1000 Genomes disagree. Variant quality information is also shown in the side tab, which provides the allele ratio and total read depth. Furthermore, you can examine the Phred quality score under the classification field, which measures the quality of the allele generated in the variant caller.
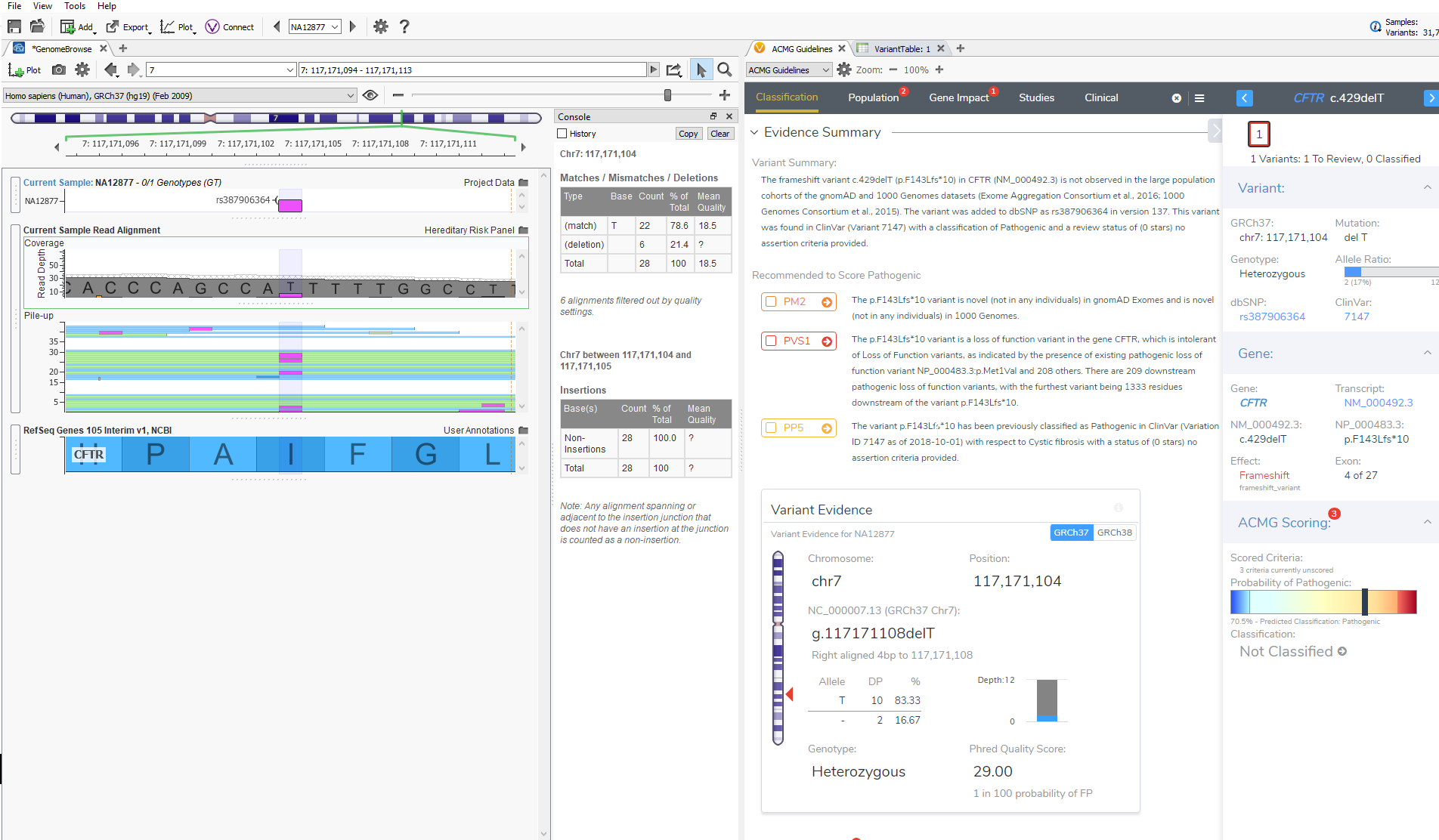
The last feature that will be discussed is GenomeBrowse. GenomeBrowse, as shown below, will now automatically display the variant that you are currently evaluating in VSClinical. In GenomeBrowse you can display the coverage and pile-up information, genotypes, and multiple annotation sources all in one view. This feature now saves the user time in having to find the variant in the variant table and will further aid in the interpretation of variants being evaluated according to the ACMG guidelines.
Taken together, VSClinical provides multiple resources to help ensure best practices when evaluating variants with the ACMG guidelines. Specifically, users can implement the Evidence, Comment and Caveat features to add validation to their evaluation, and these components can serve as a repository for future recollection. Additionally, quality details and depth of coverage are quickly assessed and interpreted to eliminate false positive variants. Hopefully, this blog helps alleviate the complexity of the ACMG criteria and provides resources for interpreting the genetic evidence. The final blog of this series focuses on our new report template that automatically pulls in your interpretation from VSClinical. Continue this series by reading it here.